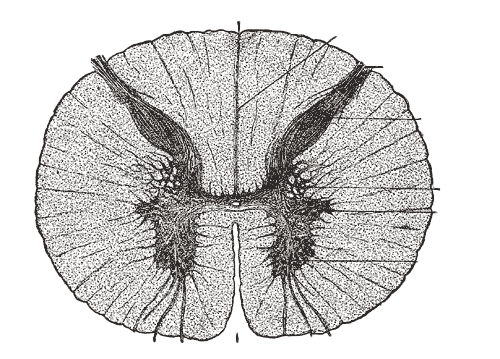
Analyze gene expression in the human spinal cord
Read the preprint by Gautier, Blum et al. 2023 here
Click a neuronal or glial spinal cord population below to analyze gene expression across diverse cell types, powered by cellxgene.
Motor neurons are stoichiometrically rare but functionally essential—they are the bridge that links the brain and body. Explore by clicking a cell type!
Click to analyze the whole dataset at once (DE analysis disabled)In-depth analysis of alpha (extrafusal muscle fibers) and gamma (intrafusal muscle fibers) motor neurons

Read the preprint by Gautier, Blum et al. 2023 here
