Human Motor Neuron Data
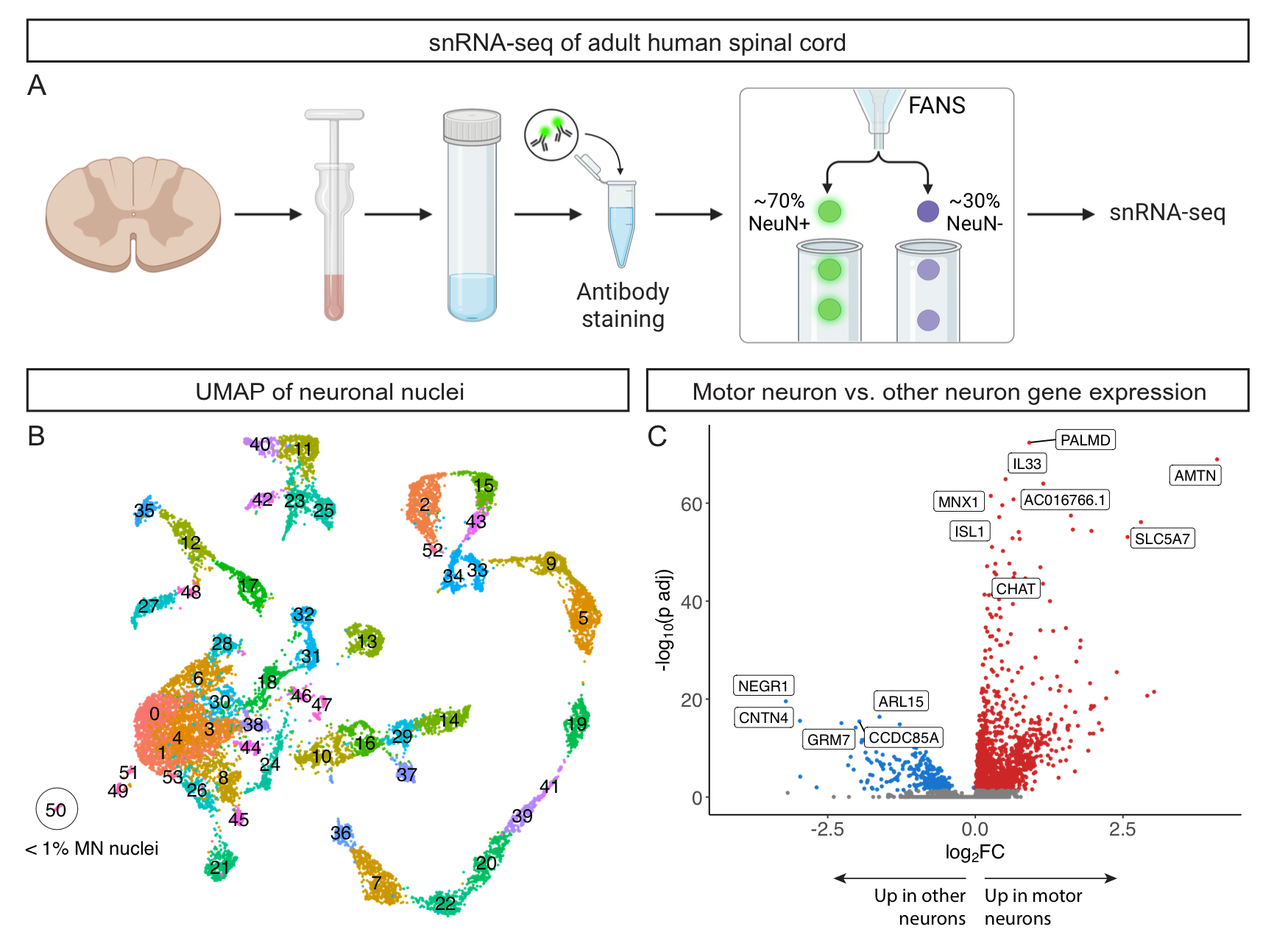
A recent paper detailing the transcriptomic diversity of the spinal cord made suggested that motor neurons were more abundant than expected, diverged highly from mouse motor neurons, and were heavily enriched for cytoskeletal transcripts implicated in ALS. Here we report contradictory results--that human motor neurons are rare in the spinal cord, can be divided into known subtypes, and share a common signature with mouse motor neurons. We further re-analyzed their data and provide evidence that their conclusions about motor neurons are the result of a technical artifact in the data. Please see read more on biorxiv here. We have made all of our sequencing data available on GEO, and all R objects are available for download from this site in the "More Information" tab. All analysis code is available on our GitHub repository
Read MoreHuman Motor Neuron Data

A recent paper detailing the transcriptomic diversity of the spinal cord made suggested that motor neurons were more abundant than expected, diverged highly from mouse motor neurons, and were heavily enriched for cytoskeletal transcripts implicated in ALS. Here we report contradictory results--that human motor neurons are rare in the spinal cord, can be divided into known subtypes, and share a common signature with mouse motor neurons. We further re-analyzed their data and provide evidence that their conclusions about motor neurons are the result of a technical artifact in the data. Please see read more on biorxiv here. We have made all of our sequencing data available on GEO, and all R objects are available for download from this site in the "More Information" tab. All analysis code is available on our GitHub repository
Read MoreHuman Motor Neuron Data

A recent paper detailing the transcriptomic diversity of the spinal cord made suggested that motor neurons were more abundant than expected, diverged highly from mouse motor neurons, and were heavily enriched for cytoskeletal transcripts implicated in ALS. Here we report contradictory results--that human motor neurons are rare in the spinal cord, can be divided into known subtypes, and share a common signature with mouse motor neurons. We further re-analyzed their data and provide evidence that their conclusions about motor neurons are the result of a technical artifact in the data. Please see read more on biorxiv here. We have made all of our sequencing data available on GEO, and all R objects are available for download from this site in the "More Information" tab. All analysis code is available on our GitHub repository
Read MorePichon Lab Paper is out in Nature Comm!
We are pleased to announce that the Le Pichon lab has recently published their single-nucleus cholinergic sequencing data in Nature Communications! Their data is featured on this site, and available to download as an integrated resource along with our (Gitler Lab) data. Big congratulations to Alkaslasi et al. for a fantastic and impactful paper! Read it here!
Read More